Publication Spotlights
Antigen-agnostic identification of poxvirus broadly neutralizing antibodies targeting OPG153
Paciello I, Rundlet EJ, Zhou L, Realini G, Perrone F, Pierleoni G, Troisi M, Postal J, Guivel-Benhassine F, Porrot F, Mullins CM, Moschese D, Cossu MV, Sabaini F, Accordini S, Gianesini N, Mantovani RP, Panza F, Fabbiani M, Tumbarello M, Antinori S, Castilletti C, Montagnani F, Schwartz O, Rappuoli R, McLellan JS, Andreano E
Sci Transl Med. 2025 Dec 10;17(828):eaeb3840. PMID: 41370398
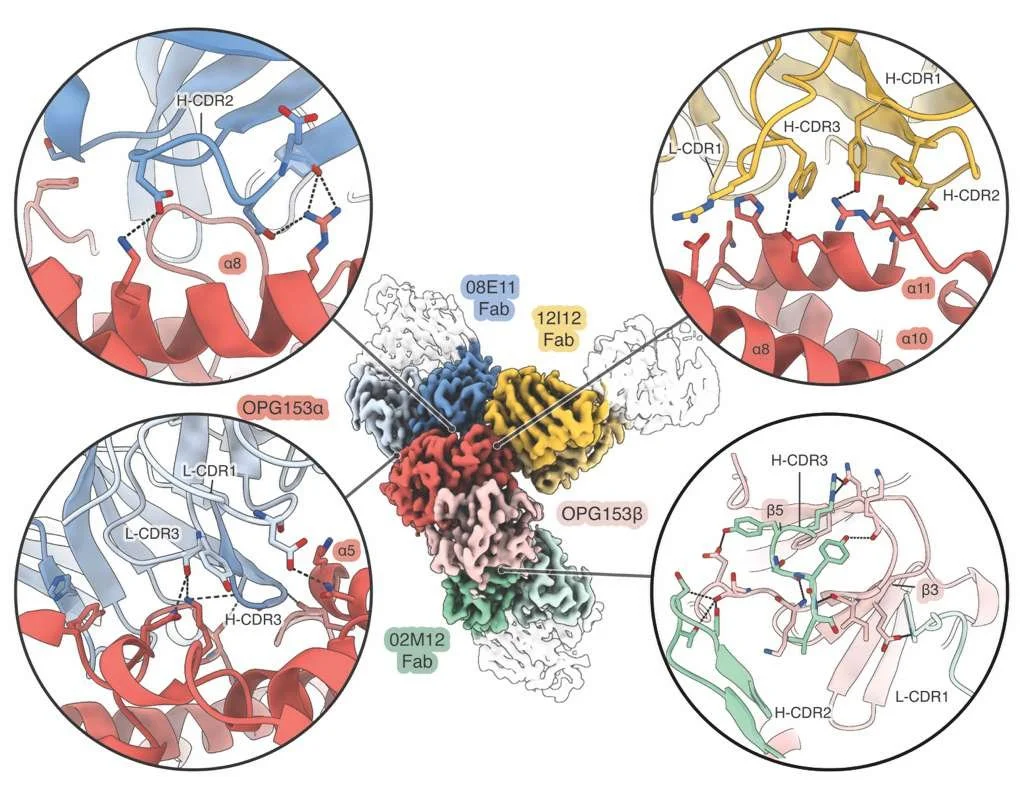
Recurrent mpox outbreaks caused by the monkeypox virus (MPXV) have prompted the World Health Organization to declare a Public Health Emergency of International Concern and have stimulated the development of medical interventions. Here, we used antigen-agnostic identification of MPXV-neutralizing monoclonal antibodies from convalescent or vaccinated people, AlphaFold 3-based predictive modeling, and cryo-electron microscopy to identify the protein encoded by orthopoxviral gene (OPG) 153 (MPXV A28) as a target of broadly neutralizing antibodies. OPG153-targeting antibodies neutralized MPXV clades Ib and IIb and vaccinia virus (VACV) and cross-reacted with OPG153 orthologs from cowpox and variola viruses. Immunization with MPXV OPG153 elicited a potent neutralizing antibody response against MPXV and VACV in mice, substantiating OPG153 as both a promising vaccine antigen and a potent target for preventive and therapeutic antibodies.
Structural basis for neutralizing antibody binding to pertussis toxin
Goldsmith JA, Nguyen AW, Wilen RE, Wijagkanalan W, McLellan JS, Maynard JA.
Proc Natl Acad Sci USA. 2025 Apr 8;122(14):e2419457122. PMID: 40172968
Pertussis toxin (PT) is a key protective antigen in vaccine- and natural immunity-mediated protection from Bordetella pertussis infection. Despite its importance, no PT-neutralizing epitopes have been characterized structurally. To define neutralizing epitopes and identify key structural elements to preserve during PT antigen design, we determined a 3.6 Å cryoelectron microscopy structure of genetically detoxified PT (PTg) bound to hu11E6 and hu1B7, two potently neutralizing anti-PT antibodies with complementary mechanisms: disruption of toxin adhesion to cells and intracellular activities, respectively. Hu11E6 binds the paralogous S2 and S3 subunits of PTg via a conserved epitope but surprisingly did not span the previously identified sialic acid-binding site implicated in toxin adhesion. Hu11E6 specifically prevented PTg binding to sialylated N-glycans and a sialylated model receptor, as demonstrated by high-throughput glycan array analysis and ELISA, while a T cell activation assay showed that it blocks PTg mitogenic activities to define its neutralizing mechanism. Hu1B7 bound a quaternary epitope spanning the S1 and S5 subunits, although functional studies of hu1B7 variants suggested that S5 binding is not involved in its PT neutralization mechanism. These results structurally define neutralizing epitopes on PT, providing key information for the future development of PT immunogens.
Engineering and structures of Crimean-Congo hemorrhagic fever virus glycoprotein complexes
McFadden E, Monticelli SR, Wang A, Ramamohan AR, Batchelor TG, Kuehne AI, Bakken RR, Tse AL, Chandran K, Herbert AS, McLellan JS.
Cell. 2025 Jan 23;188(2):303-315.e13. PMID: 39701101
Crimean-Congo hemorrhagic fever virus (CCHFV) is a tickborne virus that can cause severe disease in humans with case fatality rates of 10%-40%. Although structures of CCHFV glycoproteins GP38 and Gc have provided insights into viral entry and defined epitopes of neutralizing and protective antibodies, the structure of glycoprotein Gn and its interactions with GP38 and Gc have remained elusive. Here, we use structure-guided protein engineering to produce a stabilized GP38-Gn-Gc heterotrimeric glycoprotein complex (GP38-GnH-DS-Gc). A cryo-electron microscopy (cryo-EM) structure of this complex provides the molecular basis for GP38's association on the viral surface, reveals the structure of Gn, and demonstrates that GP38-Gn restrains the Gc fusion loops in the prefusion conformation, facilitated by an N-linked glycan attached to Gn. Immunization with GP38-GnH-DS-Gc conferred 40% protection against lethal IbAr10200 challenge in mice. These data define the architecture of a GP38-Gn-Gc protomer and provide a template for structure-guided vaccine antigen development.
Discovery and characterization of a pan-betacoronavirus S2-binding antibody
Johnson NV, Wall SC, Kramer KJ, Holt CM, Periasamy S, Richardson SI, Manamela NP, Suryadevara N, Andreano E, Paciello I, Pierleoni G, Piccini G, Huang Y, Ge P, Allen JD, Uno N, Shiakolas AR, Pilewski KA, Nargi RS, Sutton RE, Abu-Shmais AA, Parks R, Haynes BF, Carnahan RH, Crowe JE Jr, Montomoli E, Rappuoli R, Bukreyev A, Ross TM, Sautto GA, McLellan JS, Georgiev IS.
Structure. 2024 Sep 15:S0969-2126(24)00369-1. PMID: 39326419
The continued emergence of deadly human coronaviruses from animal reservoirs highlights the need for pan-coronavirus interventions for effective pandemic preparedness. Here, using linking B cell receptor to antigen specificity through sequencing (LIBRA-seq), we report a panel of 50 coronavirus antibodies isolated from human B cells. Of these, 54043-5 was shown to bind the S2 subunit of spike proteins from alpha-, beta-, and deltacoronaviruses. A cryoelectron microscopy (cryo-EM) structure of 54043-5 bound to the prefusion S2 subunit of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike defined an epitope at the apex of S2 that is highly conserved among betacoronaviruses. Although non-neutralizing, 54043-5 induced Fc-dependent antiviral responses in vitro, including antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP). In murine SARS-CoV-2 challenge studies, protection against disease was observed after introduction of Leu234Ala, Leu235Ala, and Pro329Gly (LALA-PG) substitutions in the Fc region of 54043-5. Together, these data provide new insights into the protective mechanisms of non-neutralizing antibodies and define a broadly conserved epitope within the S2 subunit.
Structure-based design of a soluble human cytomegalovirus glycoprotein B antigen stabilized in a prefusion-like conformation
Sponholtz MR, Byrne PO, Lee AG, Ramamohan AR, Goldsmith JA, McCool RS, Zhou L, Johnson NV, Hsieh CL, Connors M, Karthigeyan KP, Crooks CM, Fuller AS, Campbell JD, Permar SR, Maynard JA, Yu D, Bottomley MJ, McLellan JS
Proc Natl Acad Sci USA. 2024 Sep 10;121(37):e2404250121. PMID: 39231203
Human cytomegalovirus (HCMV) glycoprotein B (gB) is a class III membrane fusion protein required for viral entry. HCMV vaccine candidates containing gB have demonstrated moderate clinical efficacy, but no HCMV vaccine has been approved. Here, we used structure-based design to identify and characterize amino acid substitutions that stabilize gB in its metastable prefusion conformation. One variant containing two engineered interprotomer disulfide bonds and two cavity-filling substitutions (gB-C7), displayed increased expression and thermostability. A 2.8 Å resolution cryoelectron microscopy structure shows that gB-C7 adopts a prefusion-like conformation, revealing additional structural elements at the membrane-distal apex. Unlike previous observations for several class I viral fusion proteins, mice immunized with postfusion or prefusion-stabilized forms of soluble gB protein displayed similar neutralizing antibody titers, here specifically against an HCMV laboratory strain on fibroblasts. Collectively, these results identify initial strategies to stabilize class III viral fusion proteins and provide tools to probe gB-directed antibody responses.
Simulation-driven design of stabilized SARS-CoV-2 spike S2 immunogens
Nuqui X, Casalino L, Zhou L, Shehata M, Wang A, Tse AL, Ojha AA, Kearns FL, Rosenfeld MA, Miller EH, Acreman CM, Ahn SH, Chandran K, McLellan JS, Amaro RE.
Nat Commun. 2024 Aug 27;15(1):7370. PMID: 39191724
The full-length prefusion-stabilized SARS-CoV-2 spike (S) is the principal antigen of COVID-19 vaccines. Vaccine efficacy has been impacted by emerging variants of concern that accumulate most of the sequence modifications in the immunodominant S1 subunit. S2, in contrast, is the most evolutionarily conserved region of the spike and can elicit broadly neutralizing and protective antibodies. Yet, S2’s usage as an alternative vaccine strategy is hampered by its general instability. Here, we use a simulation-driven approach to design S2-only immunogens stabilized in a closed prefusion conformation. Molecular simulations provide a mechanistic characterization of the S2 trimer’s opening, informing the design of tryptophan substitutions that impart kinetic and thermodynamic stabilization. Structural characterization via cryo-EM shows the molecular basis of S2 stabilization in the closed prefusion conformation. Informed by molecular simulations and corroborated by experiments, we report an engineered S2 immunogen that exhibits increased protein expression, superior thermostability, and preserved immunogenicity against sarbecoviruses.
Crimean–Congo Hemorrhagic Fever Survivors Elicit Protective Non-Neutralizing Antibodies that Target 11 Overlapping Regions on Glycoprotein GP38
Shin OS*, Monticelli SR*, Hjorth CK*, Hornet V, Doyle M, Abelson D, Kuehne AI, Wang A, Bakken RR, Mishra AK, Middlecamp M, Champney E, Stuart L, Maurer DP, Li J, Berrigan J, Barajas J, Balinandi S, Lutwama JJ, Lobel L, Zeitlin L, Walker LM, Dye JM, Chandran K, Herbert AS, Pauli NT, McLellan JS.
Cell Rep. 2024 Jul 23;43(7):114502. PMID: 39002130
Crimean-Congo hemorrhagic fever virus can cause lethal disease in humans yet there are no approved medical countermeasures. Viral glycoprotein GP38, exclusive to Nairoviridae, is a target of protective antibodies, but extensive mapping of the human antibody response to GP38 has not been previously performed. Here, we isolate 188 GP38-specific antibodies from human survivors of infection. Competition experiments show that these antibodies bind across five distinct antigenic sites, encompassing eleven overlapping regions. Additionally, we reveal structures of GP38 bound with nine of these antibodies targeting different antigenic sites. Although these GP38-specific antibodies are non-neutralizing, several display protective efficacy equal to or better than murine antibody 13G8 in two highly stringent rodent models of infection. Together, these data expand our understanding regarding this important viral protein and inform the development of broadly effective CCHFV antibody therapeutics.
Structural basis for potent neutralization of human respirovirus type 3 by protective single-domain camelid antibodies
Johnson NV, van Scherpenzeel RC, Bakkers MJG, Ramamohan AR, van Overveld D, Le L, Langedijk JPM, Kolkman JA, McLellan JS.
Nat Commun. 2024 Jun 27;15(1):5458. PMID: 38937429
Respirovirus 3 is a leading cause of severe acute respiratory infections in vulnerable human populations. Entry into host cells is facilitated by the attachment glycoprotein and the fusion glycoprotein (F). Because of its crucial role, F represents an attractive therapeutic target. Here, we identify 13 F-directed heavy-chain-only antibody fragments that neutralize recombinant respirovirus 3. High-resolution cryo-EM structures of antibody fragments bound to the prefusion conformation of F reveal three distinct, previously uncharacterized epitopes. All three antibody fragments bind quaternary epitopes on F, suggesting mechanisms for neutralization that may include stabilization of the prefusion conformation. Studies in cotton rats demonstrate the prophylactic efficacy of these antibody fragments in reducing viral load in the lungs and nasal passages. These data highlight the potential of heavy-chain-only antibody fragments as effective interventions against respirovirus 3 infection and identify neutralizing epitopes that can be targeted for therapeutic development.
Universal paramyxovirus vaccine design by stabilizing regions involved in structural transformation of the fusion protein
Langedijk JPM, Cox F, Johnson NV, van Overveld D, Le L, van den Hoogen W, Voorzaat R, Zahn R, van der Fits L, Juraszek J, McLellan JS, Bakkers MJG.
Nat Commun. 2024 May 31;15(1):4629. PMID: 38821950
The Paramyxoviridae family encompasses medically significant RNA viruses, including human respiroviruses 1 and 3 (RV1, RV3), and zoonotic pathogens like Nipah virus (NiV). RV3, previously known as parainfluenza type 3, for which no vaccines or antivirals have been approved, causes respiratory tract infections in vulnerable populations. The RV3 fusion (F) protein is inherently metastable and will likely require prefusion (preF) stabilization for vaccine effectiveness. Here we used structure-based design to stabilize regions involved in structural transformation to generate a preF protein vaccine antigen with high expression and stability, and which, by stabilizing the coiled-coil stem region, does not require a heterologous trimerization domain. The preF candidate induces strong neutralizing antibody responses in both female naïve and pre-exposed mice and provides protection in a cotton rat challenge model (female). Despite the evolutionary distance of paramyxovirus F proteins, their structural transformation and local regions of instability are conserved, which allows successful transfer of stabilizing substitutions to the distant preF proteins of RV1 and NiV. This work presents a successful vaccine antigen design for RV3 and provides a toolbox for future paramyxovirus vaccine design and pandemic preparedness.
Prefusion-stabilized SARS-CoV-2 S2-only antigen provides protection against SARS-CoV-2 challenge
Hsieh Ching-Lin, Leist Sarah R., Miller Emily Happy, Zhou Ling, Powers John M., Tse Alexandra L., Wang Albert, West Ande, Zweigart Mark R., Schisler Rohit K., Chandran Kartik, Baric Ralph S., McLellan Jason S.
Nat Commun. 2024 Feb 20; 15:1553. PMID: 38378768
Ever-evolving SARS-CoV-2 variants of concern (VOCs) have diminished the effectiveness of therapeutic antibodies and vaccines. Developing a coronavirus vaccine that offers a greater breadth of protection against current and future VOCs would eliminate the need to reformulate COVID-19 vaccines. Here, we rationally engineer the sequence-conserved S2 subunit of the SARS-CoV-2 spike protein and characterize the resulting S2-only antigens. Structural studies demonstrate that the introduction of interprotomer disulfide bonds can lock S2 in prefusion trimers, although the apex samples a continuum of conformations between open and closed states. Immunization with prefusion-stabilized S2 constructs elicits broadly neutralizing responses against several sarbecoviruses and protects female BALB/c mice from mouse-adapted SARS-CoV-2 lethal challenge and partially protects female BALB/c mice from mouse-adapted SARS-CoV lethal challenge. These engineering and immunogenicity results should inform the development of next-generation pan-coronavirus therapeutics and vaccines.
Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein elicits antibodies targeting a membrane-proximal epitope
McCool RS, Musayev M, Bush SM, Derrien-Colemyn A, Acreman CM, Wrapp D, Ruckwardt TJ, Graham BS, Mascola JR, McLellan JS.
J Virol. 2023 Sep 22:e0092923. PMID: 37737588
Respiratory syncytial virus (RSV) is the leading cause of bronchiolitis and pneumonia in infants, infecting 90% of all children by age 2. Fusion of the RSV virion with a host cell is facilitated by the fusion protein (F). Previous work showed that immunization with a stabilized RSV F antigen elicited a diverse set of neutralizing antibodies. Here, we further investigated a group of these antibodies, known as public clonotype 2 (PC2), that appeared to be more prevalent in immunized individuals as compared to naturally infected individuals. Structural and biophysical characterizations revealed that PC2 members target the novel antigenic Site VI, despite their initial classification as Site IV binders based on patch mutation studies. We similarly investigated an antibody with high sequence similarity to PC2 members that was derived from a naturally infected adult. Initial yeast display-based competition assays suggested this antibody targeted site Ø; however, our structural studies revealed this antibody instead targets Site VI. These findings suggest that either Site VI antibodies were frequently misidentified or that soluble ectodomain vaccines enhance Site VI targeting by removing steric hindrance introduced by the viral membrane. In either case, this work has meaningful implications for how we analyze the humoral response to antigens and how those antigens themselves are rationally designed.
Structural Basis for Binding of Neutralizing Antibodies to Clostridioides difficile Binary Toxin
Goldsmith JA, Dewar V, Hermand P, Blais N, McLellan JS
Bacteriol. 2023 Apr 25;205(4):e0045622. PMID: 36951574
Clostridioides difficile is a gram-positive opportunistic human pathogen that causes 15,000 deaths annually in the United States, prompting a need for vaccine development. In addition to the important toxins TcdA and TcdB, binary toxin (CDT) plays a significant role in pathogenesis of certain C. difficile ribotypes by catalyzing the ADP-ribosylation of actin in host cells. However, the mechanisms of CDT neutralization by antibodies have not been studied, limiting our understanding of key epitopes for CDT antigen design. Therefore, we isolated neutralizing monoclonal antibodies against CDT and characterized their mechanisms of neutralization structurally and biochemically. 2.5 Å- and 2.6 Å-resolution X-ray crystal structures of the antibodies BINTOXB/22 and BINTOXB/9, respectively, in complex with CDTb—the CDT subunit that forms a heptameric pore for the delivery of toxic CDTa enzyme into the host cytosol—showed that both antibodies sterically clash with adjacent protomers in the assembled heptamer. Assessment of trypsin-induced oligomerization of the purified CDTb protoxin in vitro showed that BINTOXB/22 and BINTOXB/9 prevented the assembly of di-heptamers upon prodomain cleavage. This work suggests that the CDT oligomerization process can be effectively targeted by antibodies, which will aid in the development of C. difficile vaccines and therapeutics.
Selected Publications
Byrne PO, Fisher BE, Ambrozak DR, Blade EG, Tsybovsky Y, Graham BS, McLellan JS and Loomis RJ
Structural basis for antibody recognition of vulnerable epitopes on Nipah virus F protein
Nat Commun. 2023 Mar 17;14(1):1494.
PMID: 36932063
Goldsmith JA, DiVenere AM, Maynard JA, McLellan JS
Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin
Cell Rep. 2022 Aug 16;40(7):111196.
PMID: 35977491
Wrapp D, Ye X, Ku Z, Su H, Jones HG, Wang N, Mishra AK, Freed DC, Li F, Tang A, Li L, Jaijyan DK, Zhu H, Wang D, Fu TM, Zhang N, An Z, McLellan JS
Structural basis for HCMV Pentamer recognition by neuropilin 2 and neutralizing antibodies
Sci Adv. 2022 Mar 11;8(10):eabm2546.
PMID: 35275718
Mishra AK, Hellert J, Freitas N, Guardado-Calvo P, Haouz A, Fels JM, Maurer DP, Abelson DM, Bornholdt Z, Walker LM, Chandran K, Cosset FL, McLellan JS, Rey FA
Structural basis of synergistic neutralization of Crimean-Congo hemorrhagic fever virus by human antibodies
Science. 2022 Jan 7;375(6576):104-109.
PMID: 34793197
Goldsmith JA, DiVenere AM, Maynard JA, McLellan JS
Structural basis for antibody binding to adenylate cyclase toxin reveals RTX linkers as neutralization-sensitive epitopes
PLoS Pathog. 2021 Sep 21;17(9):e1009920.
PMID: 34547035
Mukhamedova M*, Wrapp D*, Shen CH*, Gilman MSA*, Ruckwardt TJ, Schramm CA, Ault L, Chang L, Derrien-Colemyn A, Lucas SAM, Ransier A, Darko S, Phung E, Wang L, Zhang Y, Rush SA, Madan B, Stewart-Jones GBE, Costner PJ, Homan LA, Hickman SP, Berkowitz NM, Doria-Rose NA, Morabito KM, DeKosky BJ, Gaudinski MR, Chen GL, Crank MC, Misasi J, Sullivan NJ, Douek DC, Kwong PD, Graham BS, McLellan JS, Mascola JR
Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses
Immunity 2021 Apr 13;54(4):769-780.e6.
PMID: 33823129
Hsieh CL, Goldsmith JA, Schaub JM, DiVenere AM, Kuo HC, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou CW, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, & McLellan JS
Structure-based design of prefusion-stabilized SARS-CoV-2 spikes
Science 2020 369(6510):1501-1505
PMID: 32703906
Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, Roose K, van Schie L, VIB-CMB COVID-19 Response Team, Hoffmann M, Pöhlmann S, Graham BS, Callewaert N, Schepens B, Saelens X, McLellan JS
Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies
Cell 2020 81(6):1436-1441
PMID: 32375025
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
Science 2020 367(6483):1260-1263
PMID: 32075877
Gilman MSA, Liu C, Fung A, Behera I, Jordan P, Rigaux P, Ysebaert N, Tcherniuk S, Sourimant J, Eléouët J-F, Sutto-Ortiz P, Decroly E, Roymans D, Jin Z, McLellan JS
Structure of the respiratory syncytial virus polymerase complex
Cell 2019 179(1):193-204. Full text
PMID: 31495574
Wang N, Rosen O, Wang L, Turner HL, Stevens LJ, Corbett KS, Bowman CA, Pallesen J, Shi W, Zhang Y, Leung K, Kirchdoerfer RN, Becker MM, Denison MR, Chappell JD, Ward AB, Graham BS, McLellan JS
Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD
Cell Rep. 2019 28(13):3395-3405. Full text PDF
PMID: 31553909
Gilman MSA, Furmanova-Hollenstein P, Pascual G, van ‘t Wout AB, Langedijk JPM, McLellan JS
Transient opening of trimeric prefusion RSV F proteins
Nat Commun. 2019 10(1):2105. Full text PDF
PMID: 31068578
Jones HG, Ritschel T, Pascual G, Brakenhoff JPJ, Keogh E, Furmanova-Hollenstein P, Lanckacker E, Wadia JS, Gilman MSA, Williamson RA, Roymans D, van 't Wout AB, Langedijk JP, McLellan JS
Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies
PLoS Pathog. 2018 14(3):e1006935.
PMID: 29509814
Goodwin E, Gilman MSA, Wrapp D, Chen M, Ngwuta JO, Moin SM, Bai P, Sivasubramanian A, Connor RI, Wright PF, Graham BS, McLellan JS, Walker LM
Infants infected with respiratory syncytial virus generate potent neutralizing antibodies that lack somatic hypermutation
Immunity 2018; 48(2):339-349.e5.
PMID: 29396163
Battles MB, Más V, Olmedillas E, Cano O, Vázquez M, Rodríguez L, Melero JA, McLellan JS
Structure and immunogenicity of prefusion-stabilized human metapneumovirus F glycoprotein
Nat Commun. 2017 Nov 16;8(1):1528.
PMID: 29142300
Tian D, Battles MB, Moin SM, Chen M, Modjarrad K, Kumar A, Kanekiyo M, Graepel KW, Taher NM, Hotard AL, Moore ML, Zhao M, Zheng ZZ, Xia NS, McLellan JS, Graham BS
Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein
Nat Commun. 2017 Nov 30;8(1):1877.
PMID: 29187732
Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong WP, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS
Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen
PNAS 2017 Aug 29;114(35):E7348-E7357.
PMID: 28807998
Rossey I, Gilman MS, Kabeche SC, Sedeyn K, Wrapp D, Kanekiyo M, Chen M, Mas V, Spitaels J, Melero JA, Graham BS, Schepens B, McLellan JS, Saelens X
Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state
Nat Commun. 2017 Feb 13;8:14158.
PMID: 28194013
Gilman MS, Castellanos CA, Chen M, Ngwuta JO, Goodwin E, Moin SM, Mas V, Melero JA, Wright PF, Graham BS, McLellan JS, Walker LM
Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors
Sci Immunol. 2016 Dec 16;1(6). pii: eaaj1879.
PMID: 28111638
Misasi J, Gilman MS, Kanekiyo M, Gui M, Cagigi A, Mulangu S, Corti D, Ledgerwood JE, Lanzavecchia A, Cunningham J, Muyembe-Tamfun JJ, Baxa U, Graham BS, Xiang Y, Sullivan NJ, McLellan JS
Structural and molecular basis for Ebola virus neutralization by protective human antibodies
Science. 2016 Mar 18;351(6279):1343-6.
PMID: 26917592
Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB
Pre-fusion structure of a human coronavirus spike protein
Nature. 2016 Mar 3;531(7592):118-21.
PMID: 26935699
Battles MB, Langedijk JP, Furmanova-Hollenstein P, Chaiwatpongsakorn S, Costello HM, Kwanten L, Vranckx L, Vink P, Jaensch S, Jonckers TH, Koul A, Arnoult E, Peeples ME, Roymans D, McLellan JS
Molecular mechanism of respiratory syncytial virus fusion inhibitors
Nat Chem Biol. 2016 Feb;12(2):87-93.
PMID: 26641933
Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJ, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, Langedijk JP
A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism
Nat Commun. 2015 Sep 3;6:8143
PMID: 26333350
Gilman MS, Moin SM, Mas V, Chen M, Patel NK, Kramer K, Zhu Q, Kabeche SC, Kumar A, Palomo C, Beaumont T, Baxa U, Ulbrandt ND, Melero JA, Graham BS, McLellan JS
Characterization of a prefusion-specific antibody that recognizes a quaternary, cleavage-dependent epitope on the RSV fusion glycoprotein
PLoS Pathog. 2015 Jul 10; 11(7):e1005035.
PMID: 26161532